CMSDAS Pre-Exercise 5: Using the grid
Overview
Teaching: 0 min
Exercises: 60 minQuestions
Objectives
Learn how to find CMS data on the grid
Launch a MC (Monte Carlo) generation job using CRAB
Launch a MiniAOD processing job using CRAB
Questions
For this lesson, please submit your answers using Google Form 5.
Before you start
In this set of exercises, we will learn how to do a full-scale analysis using the Worldwide LHC Computing Grid This set of exercises will take considerably longer than the others. Having your storage space set up may take several days, grid jobs can take a few days to run, and there can be problems. Although the actual effort for this exercise is only a few hours, you should set aside about a week to complete these exercises. (Note: you may or may not use CRAB during CMSDAS; however, to do a real CMS analysis, you will certainly need to use CRAB and/or the storage resources at /store/user/.)
If you encounter any problems with the exercise, please reach out on Mattermost or send an email to CMSDASATLPC@fnal.gov with a detailed description of your problem. Outside of CMSDAS, you can find help at the CRAB troubleshooting twiki or the Computing Tools forum on cms-talk.
Note
This section assumes that you have an account on the LPC computers at FNAL. How to get one is explained in the setup instructions. However, those familiar with the CERN computing environment and somewhat familiar with CRAB can answer all the questions running at CERN only. For CMSDAS, we recommend using a LPC account at FNAL and making sure you have write access to T3_US_FNALLPC. For T3_US_FNALLPC, you can get your EOS area mapped to your grid certificate by following these instructions to do a CMS Storage Space Request.
Later on, you can check with your university contact for Tier 2 or Tier 3 storage area. Once you are granted the write permission to the specified site, for later analysis you can use CRAB as the below exercise but store the output to your Tier 2 or Tier 3 storage area.
AGAIN: To perform this set of exercises, an LPC account, Grid Certificate, and CMS VO membership are required. You should already have these things, but if not, follow the setup instructions.
Finding data on the grid
In this exercise, we will learn to locate CMS data on the grid. CMS data is stored around the world at various T1, T2, and T3 computing sites. We will first learn how to use the Data Aggregation Service (DAS) to locate data (not to be confused with the data analysis school in which you are partaking!).
There are two ways to use DAS to find data: through a website or via the command line. First, let’s use the DAS website. You will be asked for your grid certificate, which you should have loaded into your browser. (Also note that there may be a security warning message, which you will need to bypass.) Enter the following into the form:
dataset dataset=/DYJetsToLL_M-50_TuneCP5_13TeV-madgraphMLM-pythia8/RunIISummer20UL18NanoAODv9-106X_upgrade2018_realistic_v16_L1v1-v1/NANOAODSIM
The first “dataset” indicates that you’re searching for whole data sets; the second “dataset=/DYJets…” is a filter, returning only this specific data set. The search results should show exactly one hit, along with several links for more info.
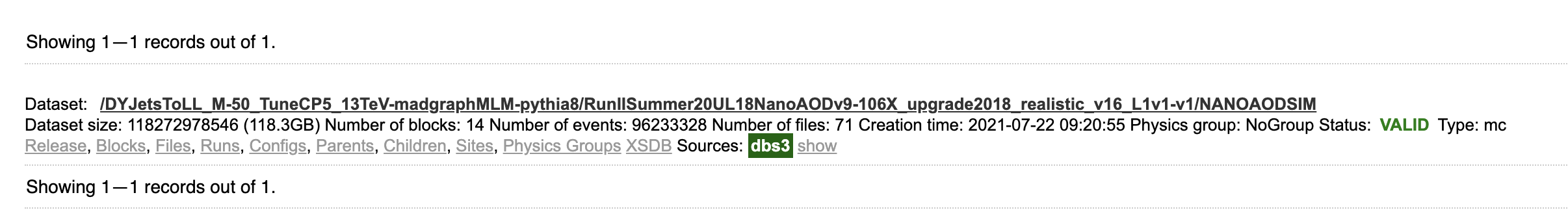
Let’s click on “Files,” which actually performs another search (you can see the exact search string in the search bar) for files in this data set. The results show 71 files, mostly around 2 GB in size with about 2 million events per file.
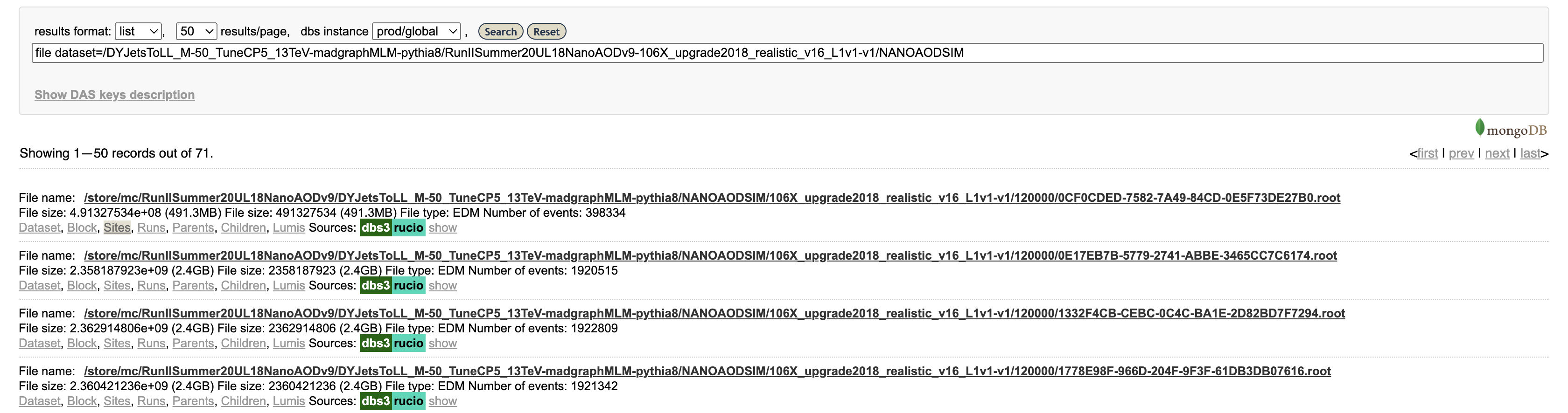
Finally, click on “Sites” for a single file. This will show the sites around the world that have this file available. This particular file is available at numerous sites, including T1_US_FNAL_Disk, T2_US_MIT, and T2_US_Purdue in the US.
More information about accessing data in the Data Aggregation Service can be found in WorkBookDataSamples.
CRAB introduction and prerequisites
Using DAS, we can locate our desired input data using, e.g., file dataset=/DYJetsToLL.... We could download all the files from the grid using xrootd. However, if you were to try this, you would quickly fill up your quota! Instead, we can use the CMS Remote Analysis Builder (CRAB). CRAB is a utility that distributes CMSSW jobs to the CMS grid (typically, but not always, using CPUs at the site where the input data is stored). The jobs will then transfer the reduced output (e.g., skimmed/slimmed ntuples or even histograms) to your /store/user/ space. CRAB is your entry point to the CMS’s significant CPU and storage resources around the world.
For general help or questions about CRAB, see the CRAB FAQ. The most recent CRAB tutorial can be found in the WorkBook under WorkBookCRABTutorial. This tutorial provides complete instructions for both beginner and expert CRAB users. We strongly recommend you to follow the CRAB tutorial after you finish these exercises.
Verify your grid certificate is OK
This exercise depends on obtaining a grid certificate and VOMS membership, but does not depend on any previous exercises.
After you’ve followed all the instructions above and installed your grid certificate, you need to verify it has all the information needed. Please install your grid certificate also on cmslpc-el8.fnal.gov
Login to cmslpc-el8.fnal.gov and initialize your proxy:
voms-proxy-init -voms cms
Then run the following command:
voms-proxy-info -all | grep -Ei "role|subject"
The response should look like this:
subject : /DC=ch/DC=cern/OU=Organic Units/OU=Users/CN=haweber/CN=713242/CN=Hannsjorg Weber/CN=2282089851
subject : /DC=ch/DC=cern/OU=Organic Units/OU=Users/CN=haweber/CN=713242/CN=Hannsjorg Weber
attribute : /cms/Role=NULL/Capability=NULL
attribute : /cms/uscms/Role=NULL/Capability=NULL
If you do not have the first attribute line listed above, you probably did not finish the VO registration in the setup. Double check this first, and otherwise reach out on Mattermost.
Question 5.1
Copy the output corresponding to the text in the output box above, using Google form 5.
Now that you have a valid grid proxy in the CMS VO, we will be able to run a command line client to check DAS to find CMS files and their information.
DAS client Question 5.2
Now let’s use the command line implementation of DAS to find some real collision data. This requires the valid CMS grid proxy. The
dasgoclientlets you query DAS just like the web interface. On cmslpc, enter the following query (along with agrepcommand to filter the result):dasgoclient -query="dataset dataset=/JetHT/Run2018C-UL2018_MiniAODv2_NanoAODv9-v1/NANOAOD" -json | grep "nevents"How many events are in this dataset? Enter the result in the Google form.
Obtain a /store/user area
This exercise pertains specifically to cmslpc. When you obtained your cmslpc account, you should have automatically been granted EOS storage at /store/user/YourUsername, however if your grid certificate was not provided or linked you may need to ensure that it is in Fermilab’s database. (As a refresher, you can review the instructions for using EOS at Using EOS at LPC.) You will configure your CRAB jobs to write output files to this space. However, it will not yet be linked to your grid certificate unless you provided it in the application form. If not linked, put in a Fermilab ServiceNow request following directions for CMS Storage Space Request. Note that this can take up to 1 business day (FNAL hours).
Setting up CRAB
Once you have a /store/user/ area connected with your grid certificate, we can start using CRAB.
In this exercise, you will generate a small MC sample yourself, and then publish it using DAS.
We will use CMSSW_13_0_13. Let’s setup a new CMSSW area for this exercise:
cd ~/nobackup/cmsdas/
scram project -n "CMSSW_13_0_13_mcgen" CMSSW_13_0_13
cd CMSSW_13_0_13_mcgen/src
cmsenv
CRAB is packaged along with CMSSW, so you should have it available now. Verify this by executing which crab; this should return something like /cvmfs/cms.cern.ch/common/crab.
Next, we will verify that your CRAB jobs have permission to write to your EOS storage. First, initialize your proxy:
voms-proxy-init -voms cms
Use the following command to check if CRAB has write access to your EOS area:
crab checkwrite --site= <site-name>
Specifically, for your cmslpc EOS storage, you should execute:
crab checkwrite --site=T3_US_FNALLPC
You should see a fair amount of output as CRAB tries several operations in your EOS storage space. At the end, hopefully you will see something like:
Will check write permission in the default location /store/user/<username>
Validating LFN /store/user/YourCERNUsername...
LFN /store/user/YourUsername is valid.
Will use `gfal-copy`, `gfal-rm` commands for checking write permissions
Will check write permission in /store/user/YourCERNUsername on site T3_US_FNALLPC
...
...
Success: Able to write in /store/user/YourCERNUsername on site T3_US_FNALLPC
Generate and publish a minimum-bias dataset with CRAB
In this exercise, we will generate a minimum bias Monte Carlo (MC) sample. A typical CMS MC job is configured with a “python fragment,” which is a small piece of python containing the necessary code to configure the generator inside CMSSW. Let’s download a recent minimum bias fragment:
cd $CMSSW_BASE/src
curl -s -k https://cms-pdmv-prod.web.cern.ch/mcm/public/restapi/requests/get_fragment/PPD-Run3Summer23GS-00003 --retry 3 --create-dirs -o Configuration/GenProduction/python/PPD-Run3Summer23GS-00003-fragment.py
The fragment contains the following lines; it mostly just sets up a special EDFilter module that runs Pythia inside CMSSW, along with a number of Pythia-specific settings:
import FWCore.ParameterSet.Config as cms
from Configuration.Generator.Pythia8CommonSettings_cfi import *
from Configuration.Generator.MCTunesRun3ECM13p6TeV.PythiaCP5Settings_cfi import *
generator = cms.EDFilter("Pythia8ConcurrentGeneratorFilter",
maxEventsToPrint = cms.untracked.int32(1),
pythiaPylistVerbosity = cms.untracked.int32(1),
filterEfficiency = cms.untracked.double(1.0),
pythiaHepMCVerbosity = cms.untracked.bool(False),
comEnergy = cms.double(13600.),
PythiaParameters = cms.PSet(
pythia8CommonSettingsBlock,
pythia8CP5SettingsBlock,
processParameters = cms.vstring(
'SoftQCD:inelastic = on'
),
parameterSets = cms.vstring('pythia8CommonSettings',
'pythia8CP5Settings',
'processParameters',
)
)
)
Next, we need to turn the fragment into a full-fledged CMSSW configuration file. We can do this with the helpful cmsDriver.py utility. Execute the following:
cd $CMSSW_BASE/src
scram b
cmsDriver.py Configuration/GenProduction/python/PPD-Run3Summer23GS-00003-fragment.py --python_filename cmsdas_minbias_cfg.py --eventcontent NANOAODGEN --step GEN,NANOGEN --datatier NANOAODSIM --customise Configuration/DataProcessing/Utils.addMonitoring --fileout file:cmsdas_minbias_nanogen.root --conditions 130X_mcRun3_2023_realistic_forMB_v1 --beamspot Realistic25ns13p6TeVEarly2023Collision --geometry DB:Extended --era Run3_2023 --no_exec --mc -n 20
Note: the scram b is necessary for CMSSW to be aware of the newly downloaded fragment file. This should have created a new file, cmsdas_minbias_cfg.py. Look over this file, and appreciate how much effort cmsDriver.py has saved us in creating this file automatically! This specific job will run 20 events through the GEN step (i.e. launching Pythia to generate truth-level events) and the NANOGEN step (which just dumps the truth particle info to a NanoAOD file); for simplicity, we are skipping the detector simulation.
Generating MC events locally
We want to test this Configuration file locally for a small number of events before we submit to CRAB for massive generation. To test this file, we can run
cmsRun cmsdas_minbias_cfg.py
This job will take a few minutes to run; you will see quite a lot of output from Pythia as it generates events. Once it finishes, you should have a brand new NANOGEN file, cmsdas_minbias_nanogen.root.
Question 5.3
What is the file size of
cmsdas_minbias_nanogen.root? Usels -lh.
Generate and publish MC dataset using CRAB
We usually need millions of MC events for a CMS analysis. The collaboration produces a lot of samples centrally, but you will occasionally need to make new samples for your analysis, or test MC. In this example, we will use CRAB to launch many minimum bias production jobs on the grid.
CRAB is configured using python configuration files (once again!). The complete documentation for these configuration files is here: CRAB3ConfigurationFile. For now, we have prepared a CRAB config for you; download it to cmslpc using:
cd $CMSSW_BASE/src
wget fnallpc.github.io/cms-das-pre-exercises/code/cmsdas_mc_crab.py
Below you also find the file:
Show/Hide
from WMCore.Configuration import Configuration config = Configuration() config.section_("General") config.General.requestName = 'cmsdas_minbias_test0' config.General.workArea = 'crabsubmit' config.section_("JobType") config.JobType.pluginName = 'PrivateMC' config.JobType.psetName = 'cmsdas_minbias_cfg.py' config.JobType.allowUndistributedCMSSW = True config.section_("Data") config.Data.outputPrimaryDataset = 'MinBias' config.Data.splitting = 'EventBased' config.Data.unitsPerJob = 20 NJOBS = 10 # This is not a configuration parameter, but an auxiliary variable that we use in the next line. config.Data.totalUnits = config.Data.unitsPerJob * NJOBS config.Data.publication = True config.Data.outputDatasetTag = 'MinBias_TuneCP5_13p6TeV-pythia8_cmsdas2025_test0' config.section_("Site") config.Site.storageSite = 'T3_US_FNALLPC'
To submit, simply execute the following:
crab submit -c cmsdas_mc_crab.py
(For the detail of the crab command, see CRABCommands.) You might be requested to enter your grid certificate password. You should get an output similar to this:
Will use CRAB configuration file cmsdas_mc_crab.py
Enter GRID pass phrase for this identity:
Importing CMSSW configuration cmsdas_minbias_cfg.py
Finished importing CMSSW configuration cmsdas_minbias_cfg.py
Sending the request to the server at cmsweb.cern.ch
Success: Your task has been delivered to the prod CRAB3 server.
Task name: 231110_212908:dryu_crab_cmsdas_minbias_test0
Project dir: crabsubmit/crab_cmsdas_minbias_test0
Please use ' crab status -d crabsubmit/crab_cmsdas_minbias_test0 ' to check how the submission process proceeds.
Log file is /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0/crab.log
Now you might notice a directory called crabsubmit is created under CMSSW_13_0_13_mcgen/src/. This directory contains all the essential metadata for your CRAB job, and is your main tool for interacting with your running jobs. Use crab status -d crabsubmit
IMPORTANT: do not modify or delete the files in this directory until you are 100% sure your job is finished! Your jobs will occasionally fail (for example, if the computing site is misconfigured, a network error occurs, the power goes out, …). You can use crab resubmit -d crabsubmit/the_job_dir to resubmit the jobs, but not if you have deleted the directory.
After you submitted the job successfully (give it a few moments), you can check the status of a task by executing the following CRAB command:
crab status -d crabsubmit/cmsdas_minbias_test0
The crab status command will produce an output containing the task name, the status of the task as a whole, the details of how many jobs are in which state (submitted, running, transfering, finished, cooloff, etc.) and the location of the CRAB log (crab.log) file. It will also print the URLs of two web pages that one can use to monitor the jobs. In summary, it should look something like this:
Show/Hide
cmslpc178:~/nobackup/cmsdas/CMSSW_13_0_13_mcgen/src --> crab status -d crabsubmit/crab_cmsdas_minbias_test0 CRAB project directory: /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0 Task name: 231110_212908:username_crab_cmsdas_minbias_test0 Grid scheduler - Task Worker: crab3@vocms0198.cern.ch - crab-prod-tw01 Status on the CRAB server: SUBMITTED Task URL to use for HELP: https://cmsweb.cern.ch/crabserver/ui/task/231110_212908%3Ausername_crab_cmsdas_minbias_test0 Dashboard monitoring URL: https://monit-grafana.cern.ch/d/cmsTMDetail/cms-task-monitoring-task-view?orgId=11&var-user=username&var-task=231110_212908%3Ausername_crab_cmsdas_minbias_test0&from=1699648148000&to=now Status on the scheduler: SUBMITTED Jobs status: idle 100.0% (10/10) No publication information available yet Log file is /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0/crab.log
Now you can take a break and have some fun. Come back after couple hours or so and check the status again.
Show/Hide
[tonjes@cmslpc101 src]$ crab status crabsubmit/cmsdas_minbias_test0 CRAB project directory: /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0 Task name: 231110_212908:username_crab_cmsdas_minbias_test0 Grid scheduler - Task Worker: crab3@vocms0122.cern.ch - crab-prod-tw01 Status on the CRAB server: SUBMITTED Task URL to use for HELP: https://cmsweb.cern.ch/crabserver/ui/task/211024_214242%3Abelt_cmsdas_minbias_test0 Dashboard monitoring URL: https://monit-grafana.cern.ch/d/cmsTMDetail/cms-task-monitoring-task-view?orgId=11&var-user=belt&var-task=211024_214242%3Abelt_cmsdas_minbias_test0&from=1635108162000&to=now Status on the scheduler: COMPLETED Jobs status: finished 100.0% (10/10) Publication status of 1 dataset(s): done 100.0% (10/10) (from CRAB internal bookkeeping in transferdb) Output dataset: /MinBias/belt-CMSDAS2021_CRAB3_MC_generation_test0-67359df6f8a0ef3c567d7c8fea38a809/USER Output dataset DAS URL: https://cmsweb.cern.ch/das/request?input=%2FMinBias%2Fbelt-CMSDAS2021_CRAB3_MC_generation_test0-67359df6f8a0ef3c567d7c8fea38a809%2FUSER&instance=prod%2Fphys03 Warning: the max jobs runtime is less than 30% of the task requested value (1250 min), please consider to request a lower value for failed jobs (allowed through crab resubmit) and/or improve the jobs splitting (e.g. config.Data.splitting = 'Automatic') in a new task. Warning: the average jobs CPU efficiency is less than 50%, please consider to improve the jobs splitting (e.g. config.Data.splitting = 'Automatic') in a new task Summary of run jobs: * Memory: 39MB min, 84MB max, 43MB ave * Runtime: 0:04:55 min, 0:45:15 max, 0:08:59 ave * CPU eff: 7% min, 73% max, 22% ave * Waste: 1:15:29 (46% of total) Log file is /uscms_data/d3/username/cmsdas/CMSSW_13_0_13_mcgen/src/crabsubmit/crab_cmsdas_minbias_test0/crab.log
Note: If you specified T3_US_FNALLPC as your output directory, CRAB will write the output to your eos area. You can see them at something like eosls /store/user/username/MinBias/MinBias_TuneCP5_13p6TeV-pythia8_cmsdas2025_test0/....
From the bottom of the output, you can see the name of the dataset and the DAS link to it. Congratulations! This is the your first CMS dataset.
There is some magic going on under the hood here. For example, if you were to simply cmsRun cmsdas_minbias_cfg.py ten times, you would get 10 identical output files. CRAB takes care of assigning each job an independent seed for the random number generator, so that each file contains unique events!
Question 5.4
What is the dataset name you published?
Processing MiniAOD with CRAB
In this exercise, we will use CRAB to run the ZPeakAnalyzer module from the previous exercise. Remember that we were using a different CMSSW release, CMSSW_13_0_10_cmsdas, so make sure to logout, login, and setup the correct release:
cd ~/nobackup/cmsdas/CMSSW_13_0_10_cmsdas/src
cmsenv
Download another CRAB configuration file,
cd $CMSSW_BASE/src
wget fnallpc.github.io/cms-das-pre-exercises/code/cmsdas_zpeak_crab.py
The configuration file looks like this:
Show/Hide
import os from WMCore.Configuration import Configuration config = Configuration() config.section_("General") config.General.requestName = 'cmsdas_zpeak_test0' config.General.workArea = 'crabsubmit' config.section_("JobType") config.JobType.pluginName = 'Analysis' config.JobType.psetName = os.path.expandvars('$CMSSW_BASE/src/MyAnalysis/LearnCMSSW/test/zpeak_cfg.py') config.JobType.allowUndistributedCMSSW = True config.section_("Data") config.Data.inputDataset = '/DoubleMuon/Run2016C-03Feb2017-v1/MINIAOD' config.Data.inputDBS = 'global' config.Data.splitting = 'LumiBased' config.Data.unitsPerJob = 50 config.Data.lumiMask = 'https://cms-service-dqmdc.web.cern.ch/CAF/certification/Collisions16/13TeV/Cert_271036-275783_13TeV_PromptReco_Collisions16_JSON.txt' config.Data.runRange = '275776-275782' config.section_("Site") config.Site.storageSite = 'T3_US_FNALLPC'
Most of this file is similar to the previous MC generation job, but there are a few key differences. The runRange parameter is used to process only a small number of runs (to save time, effort, and CO2 in this exercise). The lumiMask parameter specifies which runs and lumisections are good for analysis (each detector is responsible for certifying which lumisections are good or bad; for example, HCAL would mark data as bad if the bias voltage for the photomultipliers was incorrect).
Run CRAB
Now go through the same process for this config file. You submit it with
crab submit -c cmsdas_zpeak_crab.py
and check the status with
crab status
After a while, you should see something like below:
Show/Hide
cmslpc154:~/cmsdas/2025/test/CMSSW_13_0_10_cmsdas/src/crabsubmit --> crab status -d crab_cmsdas_zpeak_test0/ Your user certificate is going to expire in 1 days. See: https://twiki.cern.ch/twiki/bin/view/CMSPublic/WorkBookStartingGrid#ObtainingCert CRAB project directory: /uscms_data/d3/dryu/cmsdas/2025/test/CMSSW_13_0_10_cmsdas/src/crabsubmit/crab_cmsdas_zpeak_test0 Task name: 231130_123342:dryu_crab_cmsdas_zpeak_test0 Grid scheduler - Task Worker: crab3@vocms0121.cern.ch - crab-prod-tw01 Status on the CRAB server: SUBMITTED Task URL to use for HELP: https://cmsweb.cern.ch/crabserver/ui/task/231130_123342%3Adryu_crab_cmsdas_zpeak_test0 Dashboard monitoring URL: https://monit-grafana.cern.ch/d/cmsTMDetail/cms-task-monitoring-task-view?orgId=11&var-user=dryu&var-task=231130_123342%3Adryu_crab_cmsdas_zpeak_test0&from=1701344022000&to=now Status on the scheduler: COMPLETED Jobs status: finished 100.0% (31/31) Output dataset: /FakeDataset/fakefile-FakePublish-93f81f35bfff96a473e04044a2f7a529/USER No publication information available yet Warning: the max jobs runtime is less than 30% of the task requested value (1250 min), please consider to request a lower value for failed jobs (allowed through crab resubmit) and/or improve the jobs splitting (e.g. config.Data.splitting = 'Automatic') in a new task. Warning: the average jobs CPU efficiency is less than 50%, please consider to improve the jobs splitting (e.g. config.Data.splitting = 'Automatic') in a new task Summary of run jobs: * Memory: 12MB min, 783MB max, 540MB ave * Runtime: 0:01:44 min, 0:04:03 max, 0:03:52 ave * CPU eff: 6% min, 63% max, 28% ave * Waste: 3:15:10 (62% of total) Log file is /uscms_data/d3/dryu/cmsdas/2025/test/CMSSW_13_0_10_cmsdas/src/crabsubmit/crab_cmsdas_zpeak_test0/crab.log
Create reports of data analyzed
Once all jobs are finished (see crab status above), you can print a summary:
crab report
You’ll get something like this:
Show/Hide
Summary from jobs in status 'finished': Number of files processed: 64 Number of events read: 2167324 Number of events written in EDM files: 0 Number of events written in TFileService files: 0 Number of events written in other type of files: 0 Processed lumis written to processedLumis.json Summary from output datasets in DBS: Number of events: /FakeDataset/fakefile-FakePublish-93f81f35bfff96a473e04044a2f7a529/USER: 0 Additional report lumi files: Input dataset lumis (from DBS, at task submission time) written to inputDatasetLumis.json Lumis to process written to lumisToProcess.json Log file is /uscms_data/d3/dryu/cmsdas/2025/test/CMSSW_13_0_10_cmsdas/src/crabsubmit/crab_cmsdas_zpeak_test0/crab.log
Question 5.5
How many events were read? Use
crab report -d crabsubmit/crab_cmsdas_zpeak_test0.
Optional: View the reconstructed Z peak in the combined data
Using CRAB, we have analyzed a lot more events than the local job in the previous exercise. Let’s plot the fruits of our labor. We can use the hadd command to combine the histograms from the several CRAB output files:
hadd ZPeak.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_1.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_10.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_11.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_13.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_14.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_15.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_16.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_17.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_18.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_19.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_2.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_20.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_21.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_22.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_25.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_26.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_27.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_28.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_29.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_3.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_30.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_31.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_4.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_5.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_6.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_7.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_8.root root://cmseos.fnal.gov//store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/ZPeak_9.root
Note: the author of this exercise did not actually type out this command by hand! Rather, I first obtained a list of files using eosls /store/user/dryu/DoubleMuon/crab_cmsdas_zpeak_test0/231110_214958/0000/, and then used multiple cursors in Sublime Text/Visual Studio Code to turn the list into this long command. You could also generate the command programatically. Tricks like this often come in handy. Also note that the final file is less than 5GB in size. Many filesystems/networks do not do well with much larger files.
Open the new ZPeak.root file , and plot the histogram. You should have many more events than the last exercise!
Where to find more on CRAB
- CRAB Home
- CRAB FAQ
- CRAB troubleshooting guide: Steps to address the problems you experience with CRAB and how to ask for support.
- CMS Computing Tools mailing list, where to send feedback and ask support in case of jobs problem (please send to us your crab task HELP URL from crab status output).
Note also that all CMS members using the Grid subscribe to the Grid Annoucements CMS HyperNews forum. Important CRAB announcements will be announced on the CERN Computing Announcement HyperNews forum.
Last reviewed: 2024/10/28 by David Yu
Key Points
CMS data is stored around the world at various T1, T2, and T3 computing sites.
Use the Data Aggregation Service (DAS) to search for CMS data.
The CMS Remote Analysis Builder (CRAB) utility lets you launch jobs on the CMS grid.
Grid jobs run around the world, typically (but not always) using CPUs at the same site as the data.